AACR Project GENIE: Advancing Cancer Research Through Data Sharing
Steering committee chairperson Dr. Charles Sawyers answers questions about the project and the public release of the first set of genomic data
Today, the AACR Project Genomics Evidence Neoplasia Information Exchange, or GENIE for short, hit a major milestone by releasing one of the largest fully public cancer genomic data sets to date. The release also includes a limited amount of clinical data for each patient.
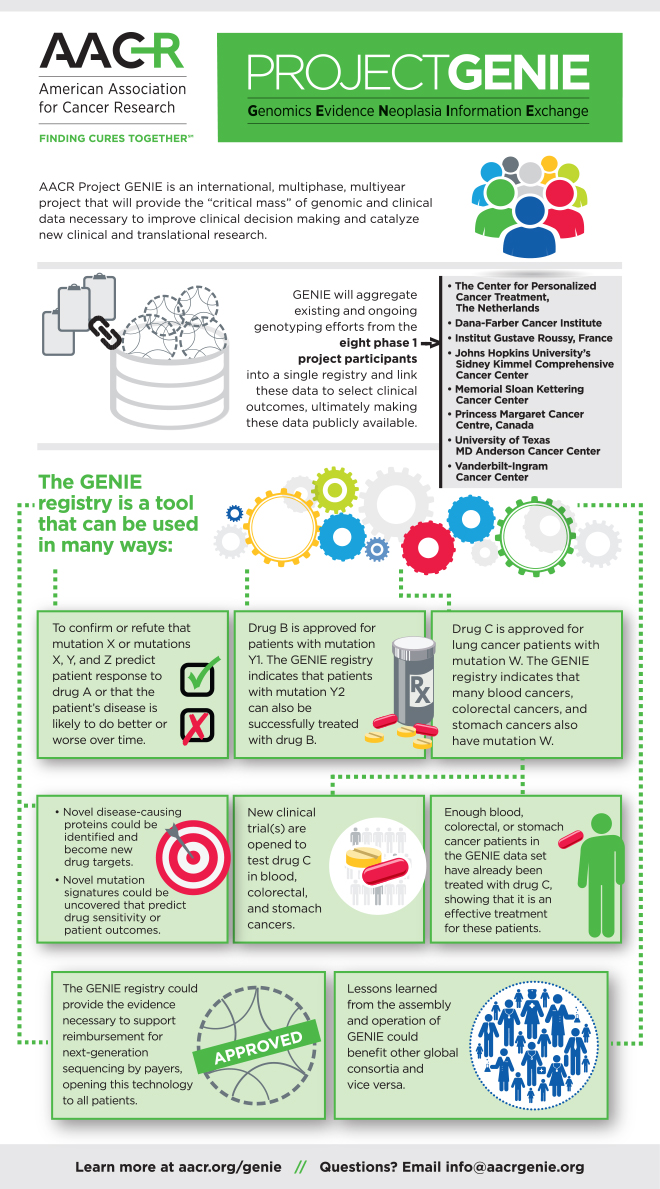
Project GENIE is a multi-phase, multi-year, international data-sharing collaboration that began with eight hospitals and cancer centers in the United States, Canada, and Europe. The project aggregates, harmonizes, and links clinical-grade, next-generation sequencing data with clinical outcomes obtained during routine medical practice from tens of thousands of cancer patients treated at these institutions. The goal of this project is to fulfill an unmet need in oncology of providing the statistical power necessary to improve clinical decision-making, particularly in the case of rare cancers and rare variants in common cancers.
We asked AACR Project GENIE Steering Committee chairperson Charles Sawyers, MD, FAACR, who is also chairperson of the Human Oncology and Pathogenesis Program at Memorial Sloan Kettering Cancer Center in New York, and a Howard Hughes Medical Institute investigator, to tell us about the new data release and the long-term goals of the project.
Question: How is AACR Project GENIE different from other data sharing projects?

Charles L. Sawyers, MD, FAACR, is the AACR Project GENIE Steering Committee Chairperson and a past president of the American Association for Cancer Research.
Answer: This is the first project I’m aware of where eight cancer centers pooled their data in a way such that the data become an open source for the cancer research community. In addition, the consortium was set up so that longitudinal outcome data can be obtained going forward.
There are already a lot of data publicly accessible from TCGA [The Cancer Genome Atlas], ICGC [International Cancer Genome Consortium], and few other sources. TCGA, for example, is a database that has comprehensive research-grade genomic data on the tumors together with some clinical data available at the time the specimens were submitted for sequencing. GENIE is different because the genomic data and clinical data are obtained from patients who are part of routine oncology practice, and the database is “living” in that additional data on these patients can be collected over time. In many of the earlier studies, the tumors had to be of a certain size or quality with stringent quality control metrics that would disqualify a lot of routine clinical samples. I would say GENIE is perhaps more of a real-world view of the breadth of frequency of different mutations across large number of cancers.
Q: What can you tell us about the data that became public today?
A: It is the first release of genomic data from roughly 19,000 patients whose tumor samples were sequenced before November 2015, meaning they are relatively recent, and they were all part of routine clinical care at these institutions. These weren’t just data from clinical trials – these were everyday patients with metastatic cancer. Basic characteristics of the patients’ tumors were also obtained at the time of biopsy. This release does not yet have information about what happened to the patients subsequently. Those data will be collected in the future.
Today’s release includes genomic data on 59 major cancer types, including data from nearly 3,000 patients with lung cancer and more than 2,000 patients each with breast cancer and colorectal cancer.
Q: Who will mostly benefit from these data immediately and how?
A: The cancer research community at large, particularly those interested in cancer genomics. While the research community has gained substantial knowledge about many common mutations that drive some types of cancers, we have a dearth of information on the rarer ones, whose significance we are slowly uncovering. The data set from GENIE will provide more robust numbers on how common such mutations are and how they are distributed across different histologies. The data set released now will not tell us whether having these rare mutations is a good prognosis or bad prognosis. That question is currently being addressed by collecting outcome data on patients with these rare mutations. We hope to complete the first two mutation-specific projects, both on rare mutations in breast cancer, in time for presentation at the AACR Annual Meeting 2017.
A: There are certain basic analyses one can run on the cBioPortal, the portal through which the data can be accessed. The data will also be made available to the Genomic Data Commons (GDC). So the analytic tools available on GDC could be applied to analyze the data from GENIE. The commitment of the eight institutions is to update the data every three months.
Q: What are the criteria for recontacting cancer centers and collecting clinical outcomes data?
A: Clinical outcome data collection is currently driven by specific questions posed by the participating institutions. Proposals are submitted to the steering committee for approval, where the decision to proceed is made based on the importance of question, feasibility, and financial resources required to collect the data. Outcomes data are collected only when there is a consensus on a research question that we agree is worth answering, because it requires enormous work and expense to retrieve clinical data.
In the first year we have focused primarily on establishing the initial database and shoring up additional financial resources to support the consortium in the future. However, we have prioritized two research questions related to breast cancer, which we are pursuing currently. And we have commercial sponsors for both of them that offset the costs associated with data collection. As we execute on those, I anticipate more demand for longitudinal collection across a spectrum of diseases. In our next meeting this month, we will be discussing the next set of research questions. One may focus on lung cancer, to see if there are genomic predictors of response to chemotherapy or other therapies. Lung cancer is the largest disease represented in the GENIE data set.
Q: How does the project protect patients’ privacy?
A: The only information that is accessible from the data set is the information on genomic mutation. Everything else is de-identified. Many patients in the GENIE consortium provided informed consent for sequencing at their institutions. We do not release germline sequencing data [which could potentially be used, with sophisticated tools and in conjunction with other publicly available databases, to re-identify patients]. All users of GENIE data have to agree to a set of principles, including not trying to identify a patient. This is an honor system that has worked well with many other non-cancer databases involving genomics and we have adopted it.
Q: Can you explain how AACR Project GENIE would help patients in the long run?
A: I see GENIE as a research tool helping clinician-scientists get access to large population cohorts that can help answer questions about whether certain mutations are good or bad for general prognosis or different treatments. I don’t envision an individual doctor or a patient going to GENIE today to ask “What’s the best treatment for me?” However, it is highly likely that the information that comes out of the project over the years could lead to changes in practice guidelines, which would then impact patients directly.
One of the main reasons for doing this project is to aggregate existing data sets into a larger registry that will provide more confidence through uniform annotation of outcomes and statistical power. GENIE could also be a resource for pharmaceutical companies that seek drug approvals by the FDA [U.S. Food and Drug Administration]. There are several examples of drugs that benefit patients whose tumors have a specific mutation. Quite often the next question is, “Do you need to do a randomized trial or not?” The patient appears to be benefiting from the drug, but how do we know whether the patient with that mutation might do perfectly well without that drug? In some cases, a well-designed disease registry might serve a control group, which could avoid the time and expense involved with having to conduct a randomized trial. This topic has been discussed a lot at the regulatory level (e.g., FDA), but there hasn’t been a registry with the size, robustness, and validity needed to serve the cancer community. We designed GENIE such that it can meet those criteria. This is one reason our sponsors from the pharmaceutical sector wanted to help us get GENIE up and running as fast as possible.
Q: How does this project complement the administration’s Cancer Moonshot initiative?
A: A top recommendation from the Blue Ribbon Panel (BRP) was to encourage data-sharing efforts such as this. GENIE, in many ways, speaks to what was prioritized by a group of distinguished cancer scientists and clinicians in the BRP report. I’m hopeful that the passage of the 21st Century Cures Act and the availability of funds for the Moonshot initiative will lead to federal support to underwrite some of the costs involved with curating all this information, collecting the outcomes data, managing the consortium, and allowing the consortium to expand.
Q: What plans do you have for expanding the size and scope of this project?
A: In January 2017 we will announce a call for new members through a formal, straightforward application process. We are prepared to double in size, to include at least 16 to 20 institutions. We’ve had preliminary conversations with many candidates. The initial eight participants are all extremely pleased with how this has gone so far. It was not a free ride – it involved a lot of work for the institutions, but the benefit these institutions derived has made the endeavor worthwhile for them. I’m optimistic.
Editor’s note: The genomic data and a limited amount of linked clinical data for each patient can be accessed via cBioPortal or downloaded directly from Sage Bionetworks.



